Ranx is a modern header-only C++ library for parallel algorithmic random number generation. Using block splitting on CPUs (OpenMP) and leapfrogging on GPUs (CUDA/ROCm/oneAPI), paired with distributions from TRNG library that avoid discarding values, Ranx wraps the engine+distribution into a device‑compatible functor and applies jump‑ahead/stride patterns provided by PCG generators so that, given the same seed, you get reproducible sequences independent of thread count or backend. In other words, Ranx fulfills all the necessary and sufficient conditions to play fair on all supported platforms.
Ranx 101¶
To start using Ranx in your code, you just need to include the header:
#include <ranx/random>That will also include all the engines and the distributions that come with it. Check here if you’re compiling for different backends.
Supported engines¶
Currently, Ranx includes all the generators from PCG family (variation of LCG), mainly because their discard(n) function takes to complete:
pcg32pcg32_oneseqpcg32_uniquepcg32_fast
pcg64pcg64_oneseqpcg64_uniquepcg64_fast
You can also use STL’s engines with Ranx, if they provide discard(n) member function. But they may neither perform well in parallel (their discard(n) is mostly , if any) nor play fair as the implementation can be platform-dependent (e.g. g++ vs. Visual C++)
Support for std::philox_engine will be added to Ranx in the near future.
Supported distributions¶
Ranx includes all the 32 distributions provided by TRNG library. You can also use STL’s distribution with Ranx, but again, they don’t warrant fair play as they may discard some values or have platform-dependent implementations.
1️⃣ Bernoulli distributions
trng::bernoulli_disttrng::binomial_disttrng::negative_binomial_disttrng::geometric_disttrng::hypergeometric_dist
2️⃣ Normal distributions
trng::normal_disttrng::lognormal_disttrng::cauchy_disttrng::chi_square_disttrng::correlated_normal_disttrng::logistic_disttrng::maxwell_disttrng::rayleigh_disttrng::truncated_normal_disttrng::student_t_dist
3️⃣ Uniform distributions
trng::uniform01_disttrng::uniform_disttrng::uniform_int_dist
4️⃣ Sampling distributions
trng::discrete_dist
trng::fast_discrete_dist
5️⃣ Poisson distributions
trng::poisson_disttrng::exponential_disttrng::gamma_disttrng::weibull_disttrng::extreme_value_disttrng::zero_truncated_poisson_dist
6️⃣ Miscellaneous distributions
trng::beta_disttrng::pareto_disttrng::powerlaw_disttrng::snedecor_f_disttrng::tent_disttrng::twosided_exponential_dist
Changing existing code to use Ranx¶
Let’s start with the serial code we introduced in the previous chapters and transform it to use Ranx for different parallel APIs/ecosystems. Let’s first include the necessary headers and function templates as we usually do:
// setting OpenMP headers and library required by Ranx
#pragma cling add_include_path("/usr/lib/llvm-9/include/openmp")
#pragma cling load("libomp.so.5")#include <iostream> // <-- std::cout and std::endl
#include <iomanip> // <-- std::setw()
#include <g3p/gnuplot> // <-- g3p::gnuplot
// function template to print the numbers
template <typename RandomIterator>
void print_numbers(RandomIterator first, RandomIterator last)
{ auto n = std::distance(first, last);
for (size_t i = 0; i < n; ++i)
{ if (0 == i % 10)
std::cout << '\n';
std::cout << std::setw(3) << *(first + i);
}
std::cout << '\n' << std::endl;
}
// function template to render two randograms side-by-side
template<typename Gnuplot, typename RandomIterator>
void randogram2
( const Gnuplot& gp
, RandomIterator first
, RandomIterator second
, size_t width = 200
, size_t height = 200
)
{ gp ("set term pngcairo size %d,%d", width * 2, height)
("set multiplot layout 1,2")
("unset key; unset colorbox; unset tics")
("set border lc '#333333'")
("set margins 0,0,0,0")
("set bmargin 0; set lmargin 0; set rmargin 0; set tmargin 0")
("set origin 0,0")
("set size 0.5,1")
("set xrange [0:%d]", width)
("set yrange [0:%d]", height)
("plot '-' u 1:2:3:4:5 w rgbimage");
for (size_t i = 0; i < width; ++i)
for (size_t j = 0; j < height; ++j)
{ int c = *first++;
gp << i << j << c << c << c << "\n";
}
gp.end() << "plot '-' u 1:2:3:4:5 w rgbimage\n";
for (size_t i = 0; i < width; ++i)
for (size_t j = 0; j < height; ++j)
{ int c = *second++;
gp << i << j << c << c << c << "\n";
}
gp.end() << "unset multiplot\n";
display(gp, false);
}#include <vector> // <-- std::vector
#include <random> // <-- std::t19937 and std::uniform_int_distribution
#include <algorithm> // <-- std::generate() and std::generate_n()
#include <functional> // <-- std::bind() and std::ref()
const unsigned long seed{2718281828};
const auto n{100};
std::vector<int> v(n);
std::mt19937 r(seed);
std::uniform_int_distribution<int> u(10, 99);
std::generate_n
( std::begin(v)
, n
, std::bind(u, std::ref(r))
);
print_numbers(std::begin(v), std::end(v));
34 91 80 72 79 21 77 70 25 65
66 12 95 35 30 26 68 75 67 63
63 29 13 64 36 37 97 99 62 47
85 12 49 90 83 46 43 15 77 91
17 41 97 22 67 42 64 91 54 91
69 93 28 26 31 69 90 37 56 25
90 14 18 20 14 25 20 90 51 55
74 52 82 72 29 85 51 93 13 11
42 87 87 54 93 11 13 80 12 18
35 73 31 73 25 76 36 96 23 32
#include <vector> // <-- std::vector
#include <ranx/random> // <-- ranx::generate_n(), ranx::bind(), pcg32, trng
const unsigned long seed{2718281828};
const auto n{100};
std::vector<int> v(n);
pcg32 r(seed);
trng::uniform_int_dist u(10, 99);
ranx::generate_n
( std::begin(v)
, n
, ranx::bind(u, r)
);
print_numbers(std::begin(v), std::end(v));
10 23 36 71 10 93 48 90 49 93
74 51 26 40 47 92 20 21 80 40
14 32 60 41 33 41 41 36 68 77
89 47 18 42 35 88 48 36 77 13
34 60 14 87 93 66 66 96 70 40
81 46 26 60 34 59 80 16 63 90
42 42 68 64 13 69 93 12 81 71
83 59 43 75 95 85 39 63 10 88
84 56 84 88 78 39 16 63 24 11
98 25 41 21 20 11 92 13 48 53
#include <vector> // <-- std::vector
#include <ranx/random> // <-- ranx::generate_n(), ranx::bind(), pcg32, trng
#include <thrust/device_vector.h>
const unsigned long seed{2718281828};
const auto n{100};
thrust::device_vector<int> v(n);
pcg32 r(seed);
trng::uniform_int_dist u(10, 99);
ranx::cuda::generate_n
( std::begin(v)
, n
, ranx::bind(u, r)
);
print_numbers(std::begin(v), std::end(v));
10 23 36 71 10 93 48 90 49 93
74 51 26 40 47 92 20 21 80 40
14 32 60 41 33 41 41 36 68 77
89 47 18 42 35 88 48 36 77 13
34 60 14 87 93 66 66 96 70 40
81 46 26 60 34 59 80 16 63 90
42 42 68 64 13 69 93 12 81 71
83 59 43 75 95 85 39 63 10 88
84 56 84 88 78 39 16 63 24 11
98 25 41 21 20 11 92 13 48 53
// no need for std::vector
#include <ranx/random> // <-- ranx::generate_n(), ranx::bind(), pcg32, trng
#include <oneapi/dpl/iterator>
#include <sycl/sycl.hpp>
const unsigned long seed{2718281828};
const auto n{100};
sycl::buffer<int> v(sycl::range(n));
pcg32 r(seed);
trng::uniform_int_dist u(10, 99);
ranx::oneapi::generate_n
( std::begin(v)
, n
, ranx::bind(u, r)
);
sycl::host_accessor va{v, sycl::read_only};
print_numbers(std::begin(va), std::end(va));
10 23 36 71 10 93 48 90 49 93
74 51 26 40 47 92 20 21 80 40
14 32 60 41 33 41 41 36 68 77
89 47 18 42 35 88 48 36 77 13
34 60 14 87 93 66 66 96 70 40
81 46 26 60 34 59 80 16 63 90
42 42 68 64 13 69 93 12 81 71
83 59 43 75 95 85 39 63 10 88
84 56 84 88 78 39 16 63 24 11
98 25 41 21 20 11 92 13 48 53
To cut a long story short, for the part related to Ranx, you just need to change std::generate()/std::generate_n() and std::bind() to the corresponding Ranx alternatives ranx::generate()/ranx::generate_n() and ranx::bind().
Checking if it plays fair¶
Now lets see if we can get the same randogram as the serial code, using the same seed/engine/distribution triplet for the parallel version:
const size_t w{240}, h{240}, n{w * h};
std::vector<int> parallel(n), serial(n);
pcg32 pr(seed), sr(seed); // start with the same engine and seed
trng::uniform_int_dist c(0, 255); // for rgb
// parallel version passing copy of the engine
ranx::generate_n(std::begin(parallel), n, ranx::bind(c, pr));
std::generate_n(std::begin(serial), n, std::bind(c, std::ref(sr)));
// instantiate the gnuplot
g3p::gnuplot gp;
// rendering two randograms side-by-side for comparison
randogram2(gp, std::begin(parallel), std::begin(serial), w, h);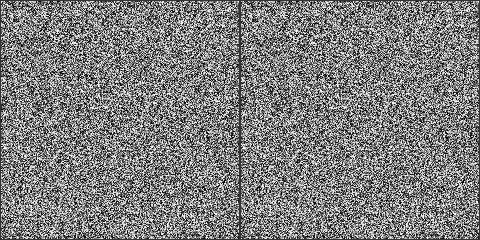
Benchmarks¶
Here’s come the moment of truth. Let’s see if our parallel versions can actually outperform the serial version. Let’s first initialize our containers:
#include <thread>
#include <execution>
#pragma cling load("libtbb.so.2")
const size_t n = 1'000'000;
std::hash<std::thread::id> hasher;
std::vector<int> s(n), rs(n), p(n), bs(n);
pcg32 r{seed}; // use the reference for the serial version%%timeit
std::generate_n
(
std::begin(s)
, n
, std::bind(u, std::ref(r))
)
;109 ms +- 20.3 ms per loop (mean +- std. dev. of 7 runs 10 loops each)
%%timeit
std::generate_n
( std::execution::par
, std::begin(rs)
, n
, [&]()
{ thread_local pcg32 r(hasher(std::this_thread::get_id()));
return u(r);
}
);30.5 ms +- 543 us per loop (mean +- std. dev. of 7 runs 10 loops each)
%%timeit
std::generate_n
( std::execution::par
, std::begin(p)
, n
, [&]()
{ thread_local pcg32 r(seed, hasher(std::this_thread::get_id()));
return u(r);
}
);41 ms +- 5.29 ms per loop (mean +- std. dev. of 7 runs 10 loops each)
%%timeit
ranx::generate_n
(
std::begin(bs)
, n
, ranx::bind(u, pcg32{seed})
)
;39.7 ms +- 5.87 ms per loop (mean +- std. dev. of 7 runs 10 loops each)